Commits on Source (253)
Showing
- .gitignore 1 addition, 0 deletions.gitignore
- README.md 62 additions, 0 deletionsREADME.md
- __init__.py 3 additions, 0 deletions__init__.py
- add_friend.py 58 additions, 0 deletionsadd_friend.py
- browse.py 19 additions, 0 deletionsbrowse.py
- compare.py 108 additions, 37 deletionscompare.py
- keras_visualize_activations/.gitignore 103 additions, 0 deletionskeras_visualize_activations/.gitignore
- keras_visualize_activations/LICENSE 201 additions, 0 deletionskeras_visualize_activations/LICENSE
- keras_visualize_activations/README.md 82 additions, 0 deletionskeras_visualize_activations/README.md
- keras_visualize_activations/__init__.py 0 additions, 0 deletionskeras_visualize_activations/__init__.py
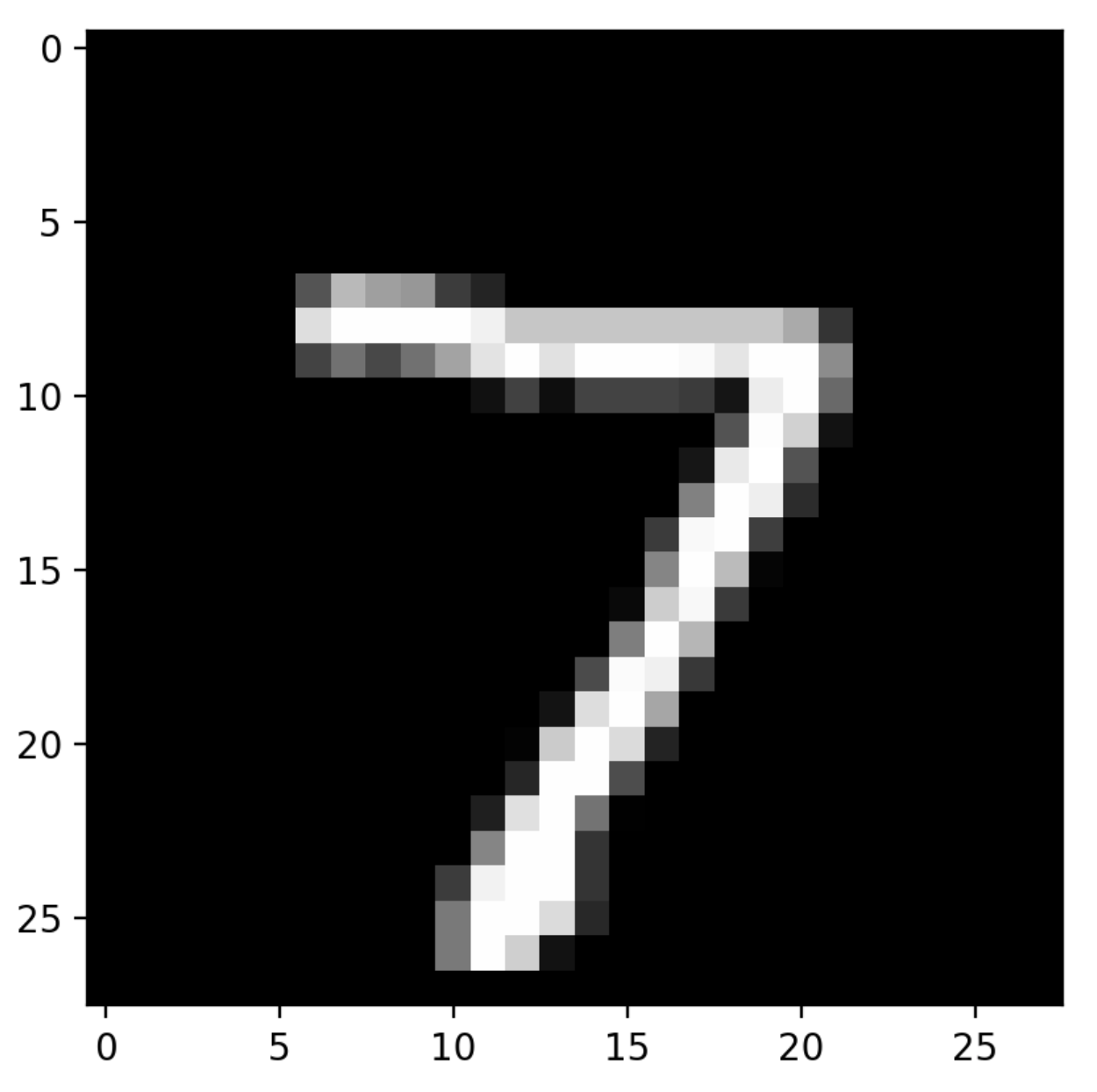
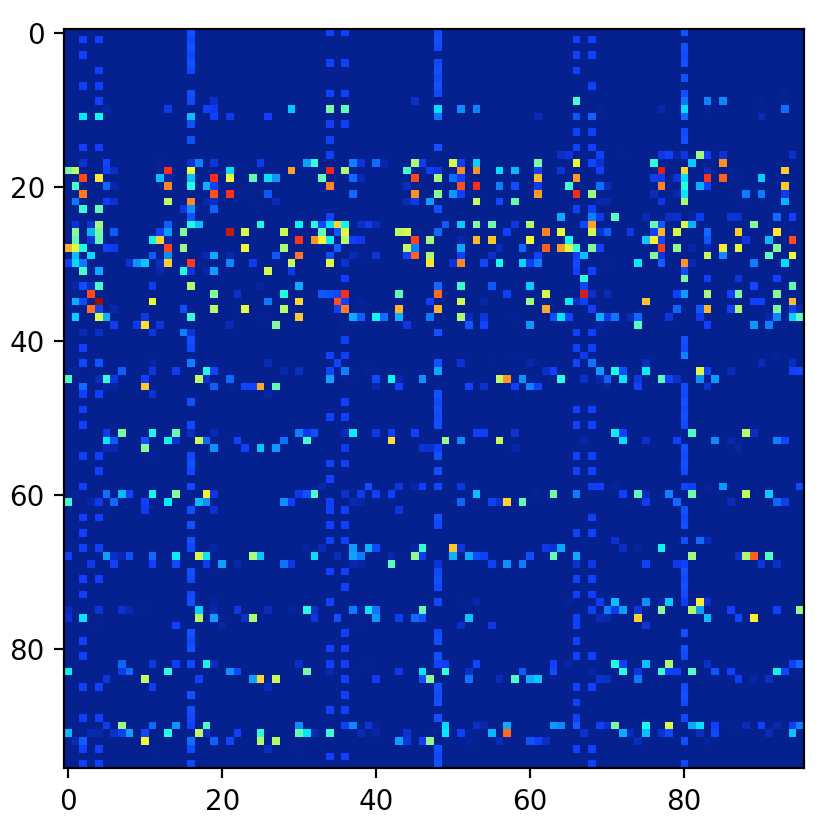
- keras_visualize_activations/assets/0.png 0 additions, 0 deletionskeras_visualize_activations/assets/0.png
- keras_visualize_activations/assets/1.png 0 additions, 0 deletionskeras_visualize_activations/assets/1.png
- keras_visualize_activations/assets/2.png 0 additions, 0 deletionskeras_visualize_activations/assets/2.png
- keras_visualize_activations/assets/3.png 0 additions, 0 deletionskeras_visualize_activations/assets/3.png
- keras_visualize_activations/checkpoints/model_09_0.990.h5 0 additions, 0 deletionskeras_visualize_activations/checkpoints/model_09_0.990.h5
- keras_visualize_activations/data.py 30 additions, 0 deletionskeras_visualize_activations/data.py
- keras_visualize_activations/model_multi_inputs_train.py 43 additions, 0 deletionskeras_visualize_activations/model_multi_inputs_train.py
- keras_visualize_activations/model_train.py 96 additions, 0 deletionskeras_visualize_activations/model_train.py
- keras_visualize_activations/read_activations.py 72 additions, 0 deletionskeras_visualize_activations/read_activations.py
- keras_visualize_activations/requirements.txt 4 additions, 0 deletionskeras_visualize_activations/requirements.txt
README.md
0 → 100644
__init__.py
0 → 100644
add_friend.py
0 → 100755
browse.py
0 → 100755
keras_visualize_activations/.gitignore
0 → 100644
keras_visualize_activations/LICENSE
0 → 100644
keras_visualize_activations/README.md
0 → 100644
keras_visualize_activations/__init__.py
0 → 100644
keras_visualize_activations/assets/0.png
0 → 100644
81.4 KiB
keras_visualize_activations/assets/1.png
0 → 100644
99.5 KiB
keras_visualize_activations/assets/2.png
0 → 100644
57.1 KiB
keras_visualize_activations/assets/3.png
0 → 100644
16.7 KiB
File added
keras_visualize_activations/data.py
0 → 100644
keras_visualize_activations/model_train.py
0 → 100644
keras_visualize_activations/requirements.txt
0 → 100644